for (element in vector) {
# do something with element
}Loop Control Statements (For Loops)
Loop Control Statements (like
forloops,whileloops) are used for performing repetitive tasks as long as a specified condition is true. It’s fundamentally about controlling the repeated execution of a code block based on a condition that is evaluated before each iteration.
for loops are powerful and general tools for streamlining repetitive tasks.
Important Note About Loops
You will interact with people who strongly discourage the use of loops in R.
This makes sense in light of the strengths of R in vectorization, but loops are excellent for beginner coders, and a very helpful fundamental to know.
They are flexible, adaptable, and effective,…
… but they are awfully slow in comparison to vectorized functions.
Though not the most efficient way to process data in R, loops get the job done. They can be translated to other programming languages, and relatively easy to understand when revisiting your code after a while.
When you interact with people who turn their nose up at certain coding practices, remember that you are not a software engineer, you are a scientist who also happens to code.
It doesn’t have to run FAST, it just has to RUN.
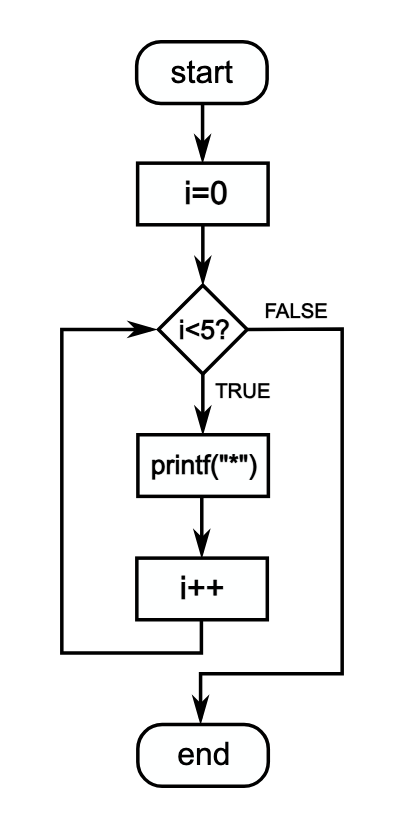
Anatomy of a For Loop
The basic structure of a for loop looks like this:
Understanding how to use for loops in R can significantly streamline your data processing tasks. This tutorial will guide you through an example using a simple data frame and demonstrate how for loops can simplify repetitive tasks.
Why Loops are Useful
Imagine we have a basic Data Frame with two columns:
df <- data.frame(val1 = c("a", "b", "c", "d", "e"),
val2 = c("d", "c", "b", "a", "e"))
df val1 val2
1 a d
2 b c
3 c b
4 d a
5 e eSuppose we want to find the row in val2 that corresponds to each row in val1.
Let’s do this individually for each value of val1
firstval <- df$val1[1]
# Viewing the first value of the first column and the whole second column
firstval[1] "a"df$val2[1] "d" "c" "b" "a" "e"# Which index of df$val2 is equal to firstval?
which(firstval == df$val2)[1] 4We repeat this process for all elements of df, manually changing the index each time.
We can create a “key” vector to map these indices:
This method is manageable for a small number of rows, but what if we had 100 rows?
This would become very impractical very quickly, which is where for loops come in.
How to Make a for loop
First, let’s understand the basic structure of a for loop:
[1] "Hey, I am i number 1"
[1] "Hey, I am i number 2"
[1] "Hey, I am i number 3"
[1] "Hey, I am i number 4"
[1] "Hey, I am i number 5"This code will print the phrase “Hey, I am i number” followed by the row of df, until it reaches the last row.
Next, modify the loop to extract and print relevant information:
for (i in 1:nrow(df)) {
print(paste("The value of column 1 row", i, "is", df$val1[i]))
keyi <- which(df$val1[i] == df$val2)
print(paste(df$val1[i], "is also found in column 2, row", keyi))
cat("\n") # Adding return to separate the phrases
}[1] "The value of column 1 row 1 is a"
[1] "a is also found in column 2, row 4"
[1] "The value of column 1 row 2 is b"
[1] "b is also found in column 2, row 3"
[1] "The value of column 1 row 3 is c"
[1] "c is also found in column 2, row 2"
[1] "The value of column 1 row 4 is d"
[1] "d is also found in column 2, row 1"
[1] "The value of column 1 row 5 is e"
[1] "e is also found in column 2, row 5"This code will print the value of val1 in each row, and the row in val2 that contains the same value until it reaches the last row.
Finally, let’s use a loop to build our key vector:
key <- vector()
for (i in 1:nrow(df)) {
keyi <- which(df$val1[i] == df$val2)
key <- c(key, keyi)
print(key) # Optional: to see the key vector being built
}[1] 4
[1] 4 3
[1] 4 3 2
[1] 4 3 2 1
[1] 4 3 2 1 5key # The final key vector[1] 4 3 2 1 5This code will build the key vector by adding the index of each row in val2 that contains the same value as the corresponding row in val1, until it reaches the last row.
The Apply Family
now, we can do this without any loops, by using the sapply() function from the apply() family of functions.
# Assuming df is a dataframe with columns val1 and val2
key <- sapply(df$val1, function(x) which(x == df$val2))
unlist(key) # it makes a list, so we need to unlist ita b c d e
4 3 2 1 5 this is a much faster technique:
result_loop <- system.time({
key <- vector()
for (i in 1:nrow(df)) {
keyi <- which(df$val1[i] == df$val2)
key <- c(key, keyi)
}
})
result_loop user system elapsed
0.002 0.000 0.002 result_vec <- system.time({
key <- sapply(df$val1, function(x) which(x == df$val2))
key_vector <- unlist(key)
})
result_vec user system elapsed
0 0 0 in this example, the vector did the same operation in less than half (0%) the amount of time as the loop, which you can imagine being a huge difference when you have a large dataset.
In day to day coding for beginning programmers, loops are easier to read and troubleshoot.
Looping Through Files
loops are also useful when you are iterating through files
for example, you would get a list of the file names in the folder. Then, you would loop over these files, reading each one and performing your operation.
# Set the path to your folder containing the files
folder_path <- "path/to/your/folder"
# Get a list of file names (assuming they are CSV files)
file_list <- list.files(path = folder_path, pattern = "\\.csv$", full.names = TRUE)
# Loop through the files
for (file in file_list) {
# Read the CSV file
data <- read.csv(file)
# Perform your operation, e.g., print the number of rows
print(paste("Number of rows in", basename(file), ":", nrow(data)))
}In this code:
-
folder_pathshould be replaced with the path to your folder. -
list.filesis used to create a list of all the.csvfiles in the folder. Thepatternargument is used to match only.csvfiles, andfull.names = TRUEreturns the full file paths. - The loop then iterates over each file name, reads the file into a data frame, and performs an operation—in this case, printing the number of rows in each file.
References and Further Reading
R is a special language and vectorisation is often most efficient option, where possible